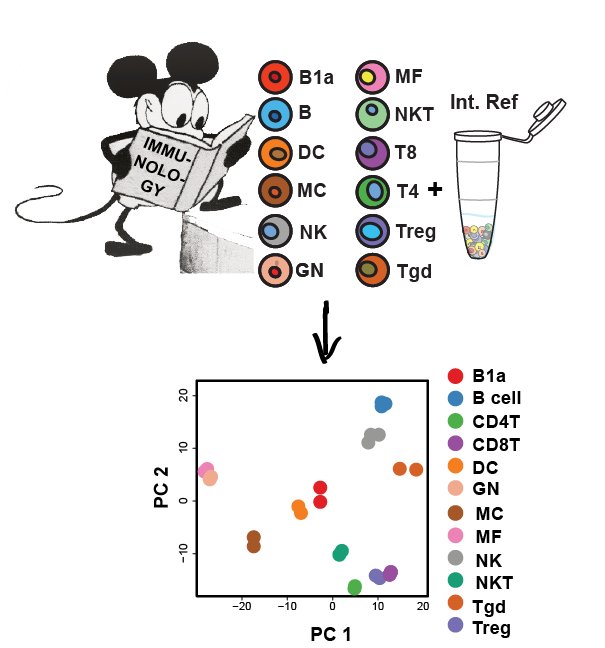The Unbearable Weight of Immesurable Molecules
One pillar of the SAM lab is technology development. There are many biological molecules, biomolecular complexes and functional phenotypes that currently have no way to be measured or characterized.
We develop and apply novel inter- and transdisciplinary approaches to study systems that have historically gone uncharacterized. The lab excels in combining genomic engineering, chemical biology, and quantitative mass spectrometry to characterize challenging protein-biomolecular interactions and understudied post-translational modifications.
Genomic Locus Proteomics
Combining CRISPR/Cas9 with proximity labeling techniques, we can now analyze the proteins associated with specific, non-repetitive loci of the mammalian genome. Instead of ChIP-seq, which looks at one protein and everywhere it occupies throughout the genome, genomic locus proteomics characterizes all the proteins at a single site in the genome.
Anti-O-GlcNAc antibodies
We have characterized new anti-O-GlcNAc antibodies in collaboration with Cell Signaling Technologies. These new antibodies enable the analysis of global O-GlcNAc patterns in cells and tissues using mass spectrometry. These new enrichment reagents will change the way O-GlcNAc signaling is studied.
Low input proteomics
Protein level measurements are important as mRNA levels do not sufficiently describe the immediate capacities of cellular responses. However, the lack of sensitivity of proteomics has historically dissuaded researchers from utilizing it for characterization efforts. We have recently developed a streamlined, quantitative proteomics workflow that is simple, accessible, and sensitive. These protocols are enabling us to ask questions about immunology and development previously unattainable by mass spectrometry.
PTM-site functional evaluation in high througput
We adopted base editors coupled to phenotypic screens to assess post-translational modification site function in high throughput.
If you have a creative idea to approach an important, unanswered biological question, we may have a spot for you….